Supercomputer Simulations of Dopamine-Derived Ligands Complexed with Cyclooxygenases
DOI:
https://doi.org/10.14529/jsfi180411Abstract
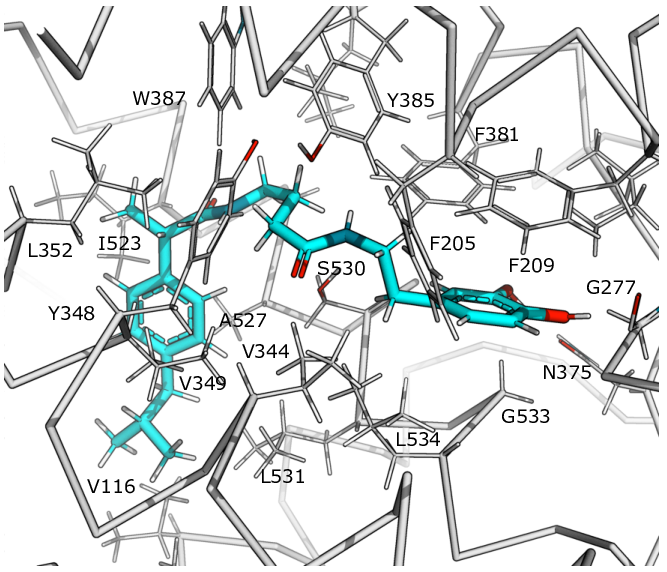
An in silico approach was adopted to identify potential cyclooxygenase inhibitors through molecular docking studies. Four potentially active molecules were generated by fusion of dopamine with ibuprofen or ketorolac derivatives. The binding mode of the considered ligands to cyclooxygenase-1 and cyclooxygenase-2 isoforms was described using Autodock Vina. Preliminary docking to full cyclooxygenase isoforms’ structures was used to determine possible binding sites for the described dopamine-derived ligands. The following more accurate docking iteration to the described binding sites was used to achieve better conformational sampling. Among the studied molecules, IBU-GABA-DA showed preferable binding to cyclooxygenase active site of cyclooxygenase-1, while IBU-DA bound to peroxidase site of cyclooxygenase-1, making these ibuprofen-comprising ligands a base for further research and design of selective cyclooxygenase-1 inhibitors. Keterolac-derived ligands KET-DA and KET-GABA-DA demonstrated binding to both cyclooxygenase isoforms at a side pocket, which does not relate to any known functional site of cyclooxygenases and needs to be further investigated.
References
Blobaum, A.L., Marnett, L.J.: Structural and functional basis of cyclooxygenase inhibition.Journal of Medicinal Chemistry 50(7), 1425–1441 (2007), DOI: 10.1021/jm06131662
Hegazy, G.H., Ali, H.I.: Design, synthesis, biological evaluation, and comparative cox1 andcox2 docking of p-substituted benzylidenamino phenyl esters of ibuprofenic and mefenamicacids. Bioorganic & Medicinal Chemistry 20(3), 1259–1270 (2012), DOI: 10.1016/j.bmc.2011.12.0303
Kummerle, A.E., da Silva, G.M.S., Sant’Anna, C.M., Barreiro, E.J., Fraga, C.A.: A proposedmolecular basis for the selective resveratrol inhibition of the PGHS-1 peroxidase activity.Bioorganic & Medicinal Chemistry 13(21), 5981–5985 (2005), DOI: 10.1016/j.bmc.2005.07.0284
Morris, G.M., Huey, R., Lindstrom, W., Sanner, M.F., Belew, R.K., Goodsell, D.S., Olson,A.J.: AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility.Journal of Computational Chemistry 30(16), 2785–2791 (2009), DOI: 10.1002/jcc.21256
O’Boyle, N.M., Banck, M., James, C.A., Morley, C., Vandermeersch, T., Hutchison, G.R.: Open babel: An open chemical toolbox. Journal of Cheminformatics 3(1), 33 (2011), DOI:10.1186/1758-2946-3-336
Reshetnikov, R.V., Stolyarova, A.V., Zalevsky, A.O., Panteleev, D.Y., Pavlova, G.V., Klinov,D.V., Golovin, A.V., Protopopova, A.D.: A coarse-grained model for DNA origami. NucleicAcids Research 46(3), 1102–1112 (2017), DOI: 10.1093/nar/gkx12627
Sadovnichy, V., Tikhonravov, A., Voevodin, V., Opanasenko, V.: “Lomonosov”: Supercom-puting at moscow state university. In: Contemporary High Performance Computing: FromPetascale toward Exascale. pp. 283–307. Chapman & Hall/CRC Computational Science, BocaRaton, United States, Boca Raton, United States (2013)
Trott, O., Olson, A.J.: Autodock vina: Improving the speed and accuracy of docking with anew scoring function, efficient optimization, and multithreading. Journal of ComputationalChemistry 31(2), 455–461 (2010), DOI: 10.1002/jcc.213349
Yoo, J.M., Park, E.S., Kim, M.R., Sok, D.E.: Inhibitory effect of n-acyl dopamines on IgE-mediated allergic response in RBL-2h3 cells. Lipids 48(4), 383–393 (2013), DOI: 10.1007/s11745-013-3758-6
Downloads
Published
How to Cite
Issue
License
Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution-Non Commercial 3.0 License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
